Sequedex classifies DNA sequences:
Who they came from and What they do
Advantages
- Starting with version 2, Sequedex is open source
- Analyzes collections of sequences in new ways, answering who is doing what? and bridging molecular to ecological scales of biology via evolution.
- Extremely fast and resource-sparing. A single core of your laptop can be ~250,000X faster than BLAST and twice as fast as uploading your data to a cloud. Sequedex tames the data deluge that has been widely noted, bringing analysis costs back into line with sequencing costs without cloud security and outage issues.
- Works for reads as short as 30 bp where other methods
fail. You don't have to distort your data through a
non-linear filter like assembly just to get results. Linear
and broad diagnostics enables
ecological management strategies
for tough problems.
- Phylogenetic signature algorithm works for novel organisms, so you're not blind to microbes that aren't similar to those in genome databases. More sensitive to distant relationships than methods based on mapping methods.
- We already built a Tree of Life for you, so your analysis doesn't have to wait on a team of bioinformaticians to finish their analysis first.
Results Gallery (clickable)
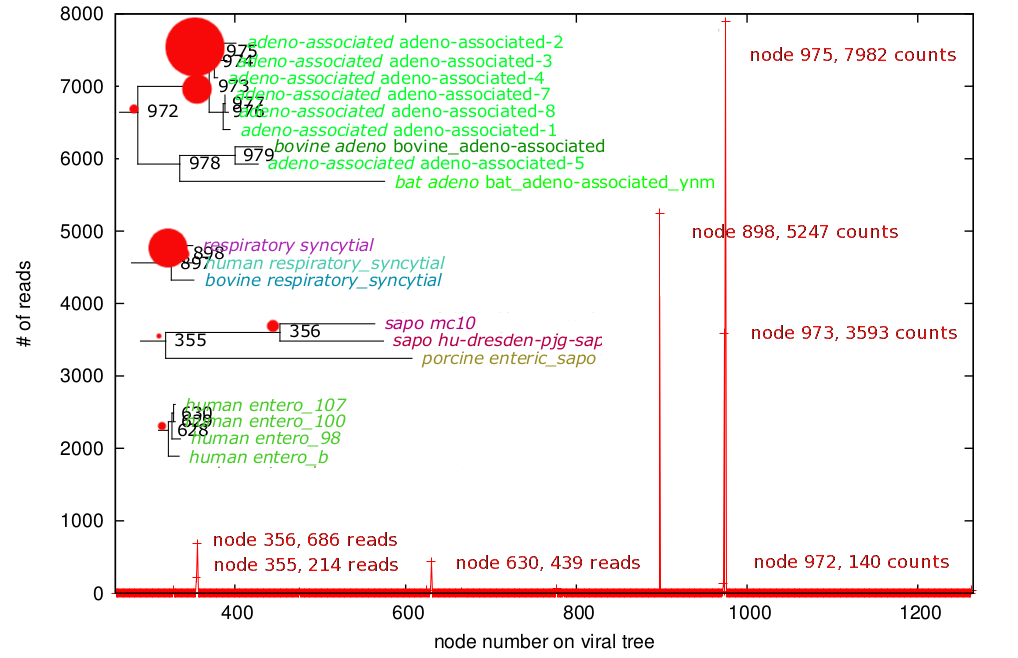
Who's there? A bunch of viruses! Viral metagenomes from a set of patients with diarrhea. It looks like the cause of their illness was an enterovirus. Data courtesy Dr. Charles Chiu, UCSF.
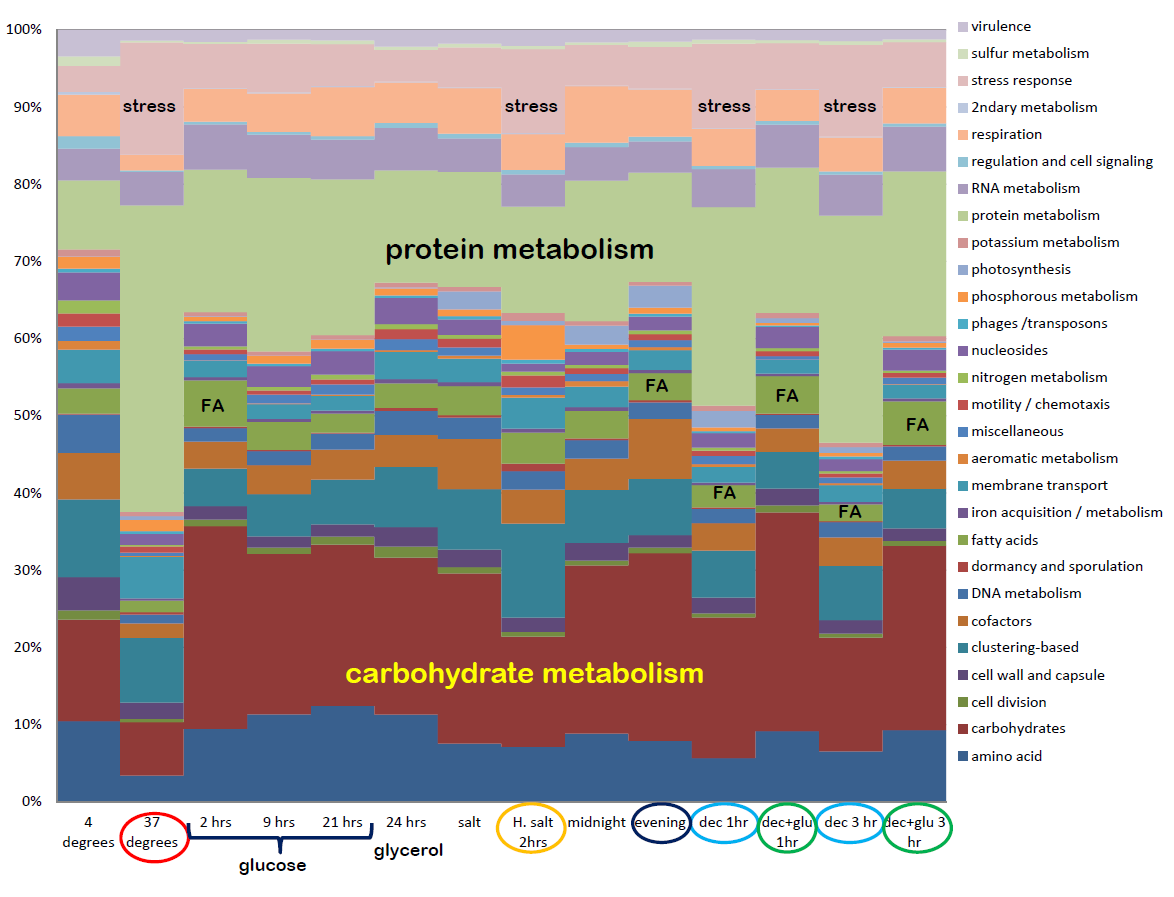
What are those algae doing? Sequedex output from a series of algal transcriptomes under various treatments. The organism under study here, Chlorella, has no reference genome or transcriptome. Data courtesy Dick Sayre, LANL.
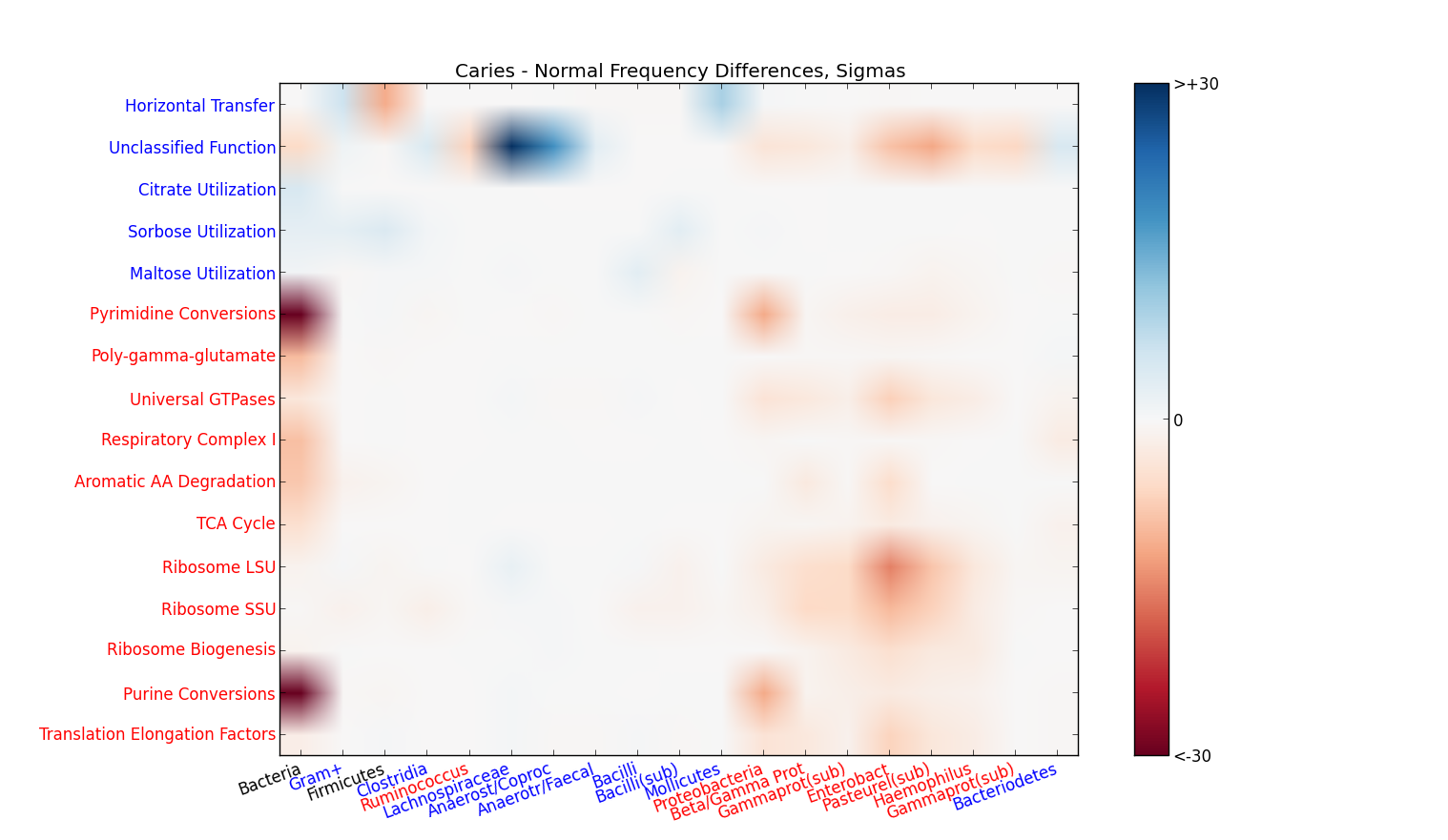
Complex communities, simple differences: People who have tooth decay show huge differences (>40 sigmas) in the frequency of genes that let bacteria live off sugars instead of nucleic acids. Blue dots are increased in caries, red is decreased. Data from NCBI SRA study SRP007831
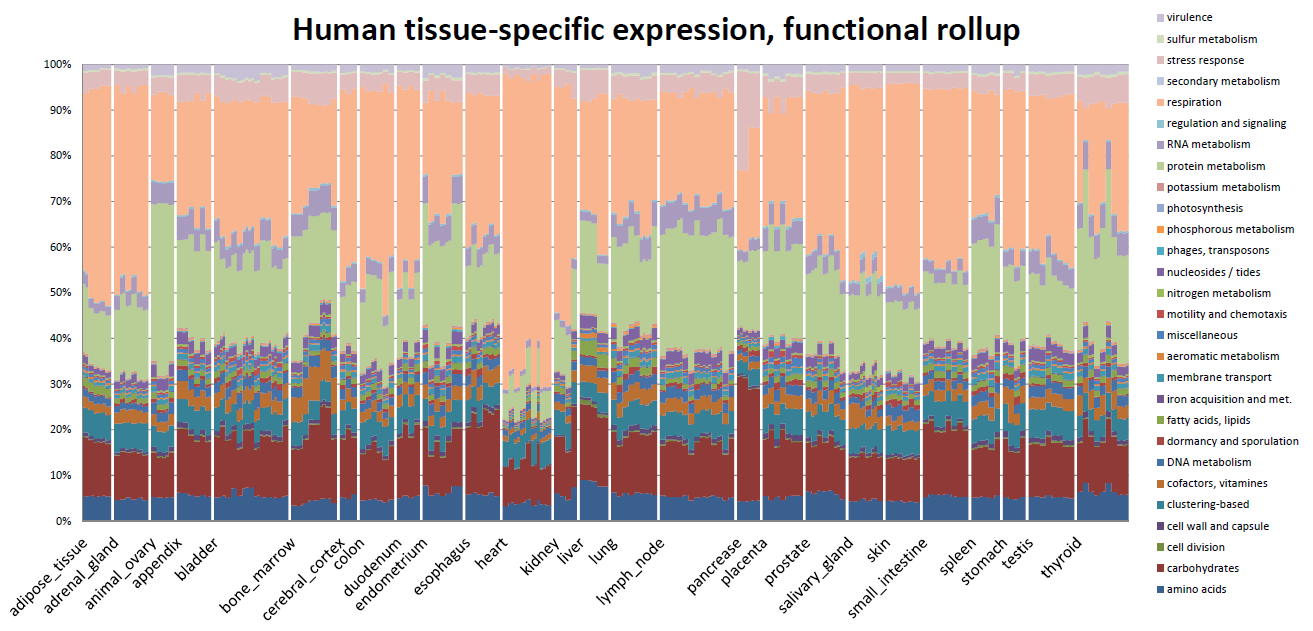
Tissue differentiation by transcriptomes. Data from the MPSS atlas of human gene expression
Applications
Potential applications for Sequedex exist anywhere analysis of biological sequences (bioinformatics) may be informative, including:
- ID of infectious diseases whether arising from bacteria, parasites, or viruses (medical, public health, biodefense).
- Characterizing gut, oral, and skin microbiomes of humans, livestock, and pets (medical, pharmaceutical, veterinary, consumer).
- Metagenomics of soils, rivers, oceans, foods, and process environments. (ecology, manufacturing).
- Transcriptome characterization (medical, pharmaceutical, agricultural, biofuels).
- Enzyme mining (chemical manufacturing, biofuels).
- Cancer genomics (medical, pharmaceutical).
- Companion diagnostics (pharmaceutical, agricultural, consumer care).